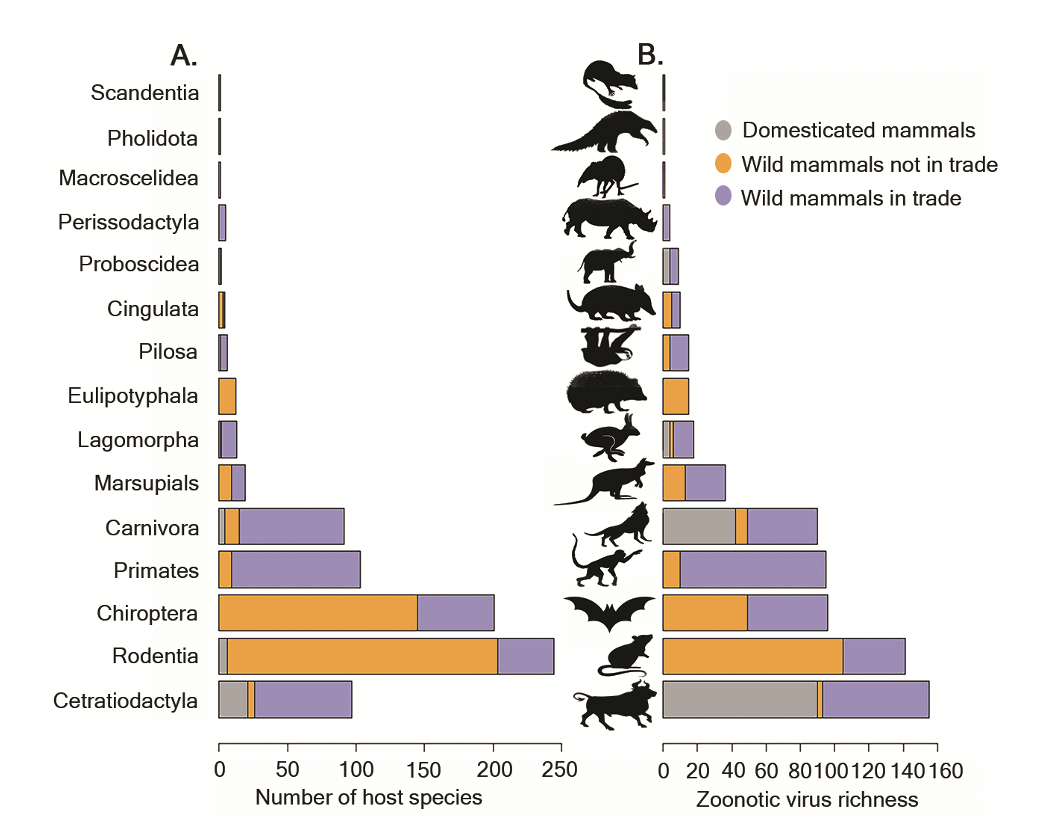
New publication presents some of the first empirical evidence showing a strong association between wildlife trade and zoonotic disease risk. In fact, scientists found that slightly more than 25 percent of mammal species found in wildlife trade host 75 percent of viruses known to be transmissible between animals and humans.
Gist
“Most new infectious diseases emerge when pathogens transfer from animals to humans,” said TNC senior scientist and first author Shivaprakash Nagaraju. “The debate around the suspected origins of the COVID-19 pandemic has resurfaced debates on the role of the wildlife trade as a potential source of emerging zoonotic diseases. Yet, there have been no studies quantitatively assessing zoonotic disease risk associated with wildlife trade. At least not until now.”
Published in Current Biology, the study combined data on mammal species hosting zoonotic viruses and data on mammals known to be in the wildlife trade, and found the traded wild mammals harbored a higher number of viruses that infect humans than domesticated and non-traded mammals. Scientists further found that traded mammals harbor distinct compositions of zoonotic viruses and different host reservoirs than non-traded and domesticated mammals.
Primates, ungulates (hoofed mammals), bats and other carnivorous mammals represent significant zoonotic disease risk as they host 132 (58 percent) of 226 known zoonotic viruses in the current wildlife trade. Looking ahead, species of bats, rodents and marsupials are likely to represent significant zoonotic disease risk in the wildlife trade of the future.
The Big Picture
Overall, the study findings strengthen the evidence that wildlife trade and zoonotic disease risks are strongly associated, and that mitigation measures should prioritize species with the highest risk of carrying zoonotic viruses.
It’s important, the authors note, to recognize the risk that a species carries zoonotic diseases is not equal for all mammal species in the wildlife trade and not all mammals in the trade host viruses that are known to be harmful to humans.
As the world looks for ways to prevent the next million-death pandemic before it starts, many global health and conservation NGOs, including TNC, are calling for investments to prevent pandemics at the source by, among other things, preventing the spillover of pathogens from animals to people.
Specifically, the groups advocate for a One Health approach that focuses on achieving positive outcomes by recognizing the interconnection between people, plants, animals and their shared environment. There is special emphasis on prioritizing support for Indigenous Peoples and local communities, as well as key tropical forest nations.
Tropical forests are linked to human health in many ways, including harboring diverse wildlife species that are reservoirs for zoonotic diseases. Deforestation in tropical forests, in particular, harms and displaces local communities and cultures, increases human-wildlife contact, and creates significant spillover risk.
The Takeaway
Curbing the sale of wildlife products and developing principles that support the sustainable and healthy trade of wildlife could be a cost-effective investment given the potential risk and consequences of zoonotic disease outbreaks.
References:
Shivaprakash, K.N., Sen, S., Paul, S., Kiesecker, J.M., Bawa, K.S., "Mammals, wildlife trade, and the next global pandemic," Current Biology, 2021, ISSN 0960-9822. https://doi.org/10.1016/j.cub.2021.06.006




Join the Discussion